API Documentation¶
All modules above listed are under the “medicaltorch” namespace.
Contents:
medicaltorch.datasets – Datasets¶
-
class
medicaltorch.datasets.MRI2DSegmentationDataset(filename_pairs, slice_axis=2, cache=True, transform=None, slice_filter_fn=None, canonical=False)[source]¶ This is a generic class for 2D (slice-wise) segmentation datasets.
Parameters: - filename_pairs – a list of tuples in the format (input filename, ground truth filename).
- slice_axis – axis to make the slicing (default axial).
- cache – if the data should be cached in memory or not.
- transform – transformations to apply.
-
class
medicaltorch.datasets.SCGMChallenge2DTest(root_dir, slice_axis=2, site_ids=None, subj_ids=None, cache=True, transform=None, slice_filter_fn=None, canonical=False)[source]¶ This is the Spinal Cord Gray Matter Challenge dataset.
Parameters: - root_dir – the directory containing the test dataset.
- site_ids – a list of site ids to filter (i.e. [1, 3]).
- subj_ids – the list of subject ids to filter.
- transform – the transformations that should be applied.
- cache – if the data should be cached in memory or not.
- slice_axis – axis to make the slicing (default axial).
Note
This dataset assumes that you only have one class in your ground truth mask (w/ 0’s and 1’s). It also doesn’t automatically resample the dataset.
See also
Prados, F., et al (2017). Spinal cord grey matter segmentation challenge. NeuroImage, 152, 312–329. https://doi.org/10.1016/j.neuroimage.2017.03.010
Challenge Website: http://cmictig.cs.ucl.ac.uk/spinal-cord-grey-matter-segmentation-challenge
-
class
medicaltorch.datasets.SCGMChallenge2DTrain(root_dir, slice_axis=2, site_ids=None, subj_ids=None, rater_ids=None, cache=True, transform=None, slice_filter_fn=None, canonical=False, labeled=True)[source]¶ This is the Spinal Cord Gray Matter Challenge dataset.
Parameters: - root_dir – the directory containing the training dataset.
- site_ids – a list of site ids to filter (i.e. [1, 3]).
- subj_ids – the list of subject ids to filter.
- rater_ids – the list of the rater ids to filter.
- transform – the transformations that should be applied.
- cache – if the data should be cached in memory or not.
- slice_axis – axis to make the slicing (default axial).
Note
This dataset assumes that you only have one class in your ground truth mask (w/ 0’s and 1’s). It also doesn’t automatically resample the dataset.
See also
Prados, F., et al (2017). Spinal cord grey matter segmentation challenge. NeuroImage, 152, 312–329. https://doi.org/10.1016/j.neuroimage.2017.03.010
Challenge Website: http://cmictig.cs.ucl.ac.uk/spinal-cord-grey-matter-segmentation-challenge
-
class
medicaltorch.datasets.SegmentationPair2D(input_filename, gt_filename, cache=True, canonical=False)[source]¶ This class is used to build 2D segmentation datasets. It represents a pair of of two data volumes (the input data and the ground truth data).
Parameters: - input_filename – the input filename (supported by nibabel).
- gt_filename – the ground-truth filename.
- cache – if the data should be cached in memory or not.
- canonical – canonical reordering of the volume axes.
-
get_pair_data()[source]¶ Return the tuble (input, ground truth) with the data content in numpy array.
medicaltorch.transforms – Transformations¶
-
class
medicaltorch.transforms.CenterCrop2D(size, labeled=True)[source]¶ Make a center crop of a specified size.
Parameters: segmentation – if it is a segmentation task. When this is True (default), the crop will also be applied to the ground truth.
-
class
medicaltorch.transforms.Normalize(mean, std)[source]¶ Normalize a tensor image with mean and standard deviation.
Parameters: - mean – mean value.
- std – standard deviation value.
medicaltorch.metrics – Metrics¶
medicaltorch.models – Models¶
-
class
medicaltorch.models.NoPoolASPP(drop_rate=0.4, bn_momentum=0.1, base_num_filters=64)[source]¶ 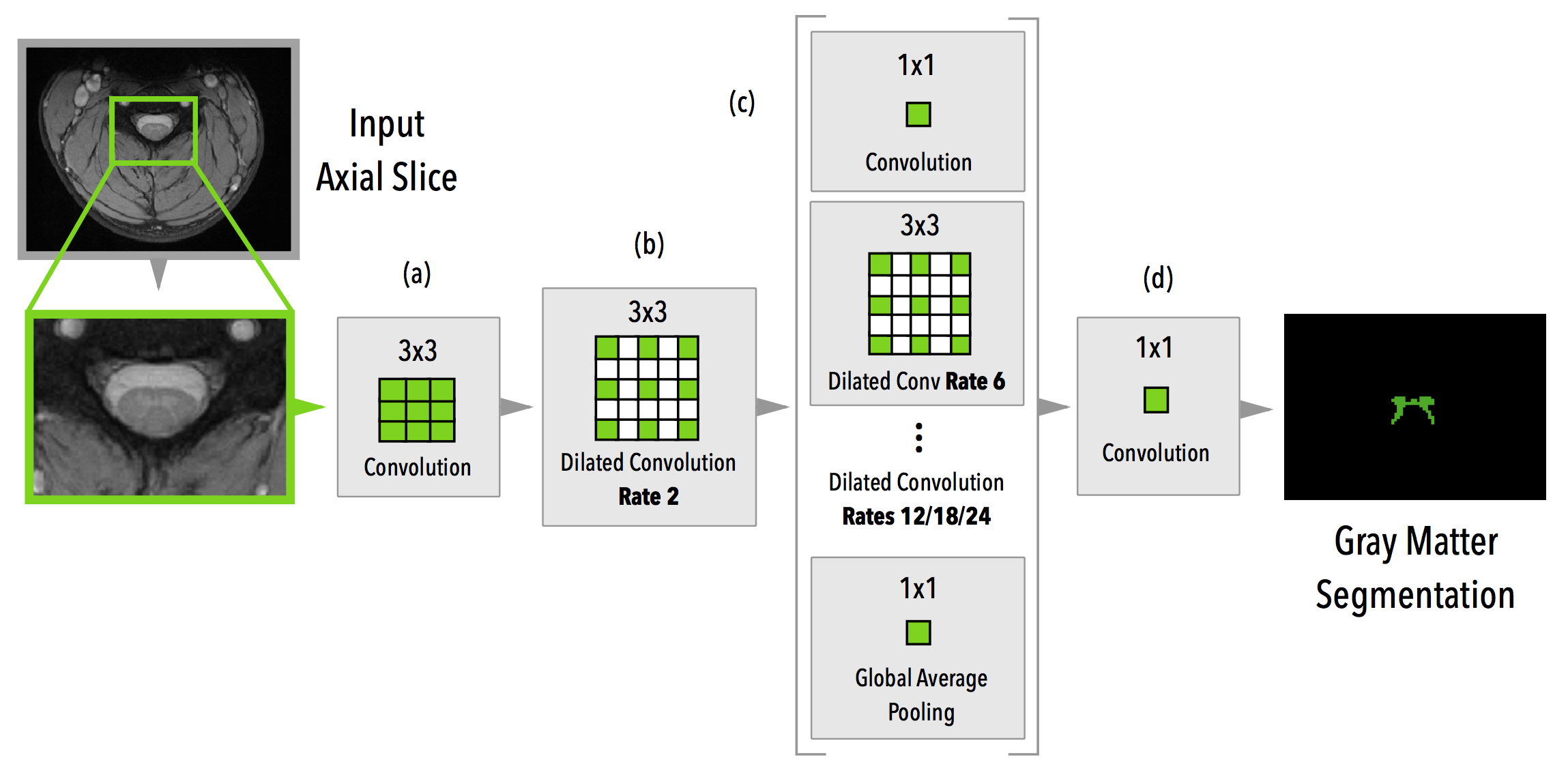
An ASPP-based model without initial pooling layers.
Parameters: - drop_rate – dropout rate.
- bn_momentum – batch normalization momentum.
See also
Perone, C. S., et al (2017). Spinal cord gray matter segmentation using deep dilated convolutions. Nature Scientific Reports link: https://www.nature.com/articles/s41598-018-24304-3
-
class
medicaltorch.models.Unet(drop_rate=0.4, bn_momentum=0.1)[source]¶ A reference U-Net model.
See also
Ronneberger, O., et al (2015). U-Net: Convolutional Networks for Biomedical Image Segmentation ArXiv link: https://arxiv.org/abs/1505.04597
